Abstract
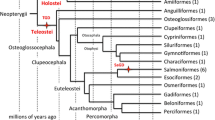
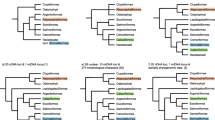
Reconstructing a well-supported phylogenetic relationship among species is a prerequisite for biological and evolutionary researches. Chondrichthyes (sharks, rays, skates and chimaeras) serves as an important clade of highly commercial, ecological, evolutionary, and conservation significance. Yet the genome-scale data of chondrichthyans are lacking, which largely restrict phylogeny, evolution, and conservation investigations of these species. Recently reported 1105 orthologous exon markers generated for ray-finned fishes are powerful tools for phylogenomics inferences. Given the relatively close relationship between cartilaginous and ray-finned fish species, we purposed to verify whether these 1105 genomic exon markers generated for ray-finned fishes were applicable for chondrichthyan phylogenomics. In this study, we collected publically available 10 genomes and 30 transcriptomes of chondrichthyans to conduct the phylogenomics analysis. Phylogenomics analyses based on the 1105 genomic exon markers and the Benchmarking Universal Single-Copy Orthologs (BUSCO) approach were performed to reconstruct phylogenetic relationships of investigated chondrichthyans. We also attempted to apply these exon markers for phylogeny of 26 jawed fish species. Our results revealed well-supported and almost identical phylogenetic relationships of chondrichthyans, confirming the applicability of genomic exon markers of ray-finned fishes in chondrichthyan phylogeny. Moreover, phylogenomics analyses of 26 jawed fish species further confirmed the application of these genomic exon markers in both cartilaginous and lobe-finned fish phylogeny. Our results highlighted the wide-ranging utility of genomic markers of ray-finned fishes in phylogenetic inference of cartilaginous and lobe-finned fish species and provided substantial and background information for the evolutionary biology and conservation of chondrichthyans and related fish species.


Similar content being viewed by others
Data availability
The genomes and transcriptomes of investigated species are available in NCBI database with accession numbers listed in supplementary Table S1. The primary information including raw aligned gene datasets and phylogenetic topologies based on two approaches were deposited in Figshare repository (https://doi.org/10.6084/m9.figshare.20291325.v2).
References
Agashe, D., & Shankar, N. (2014). The evolution of bacterial DNA base composition. Journal of Experimental Zoology. Part B, Molecular and Developmental Evolution, 322, 517–528.
Allio, R., Scornavacca, C., Nabholz, B., Clamens, A. L., Sperling, F. A., & Condamine, F. L. (2020). Whole genome shotgun phylogenomics resolves the pattern and timing of wwallowtail butterfly evolution. Systematic Biology, 69, 38–60.
Aschliman, N. C., Nishida, M., Miya, M., Inoue, J. G., Rosana, K. M., & Naylor, G. J. (2012). Body plan convergence in the evolution of skates and rays (Chondrichthyes: Batoidea). Molecular Phylogenetics and Evolution, 63, 28–42.
Betancur-R, R., Li, C., Munroe, T. A., Ballesteros, J. A., & Ortí, G. (2013). Addressing gene tree discordance and non-stationarity to resolve a multi-locus phylogeny of the flatfishes (Teleostei: Pleuronectiformes). Systematic Biology, 62, 763–785.
Capella-Gutiérrez, S., Silla-Martínez, J. M., & Gabaldón, T. (2009). trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics, 25, 1972–1973.
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution, 17, 540–552.
Clarke, S. C., McAllister, M. K., Milner-Gulland, E. J., Kirkwood, G. P., Michielsens, C. G., Agnew, D. J., et al. (2006). Global estimates of shark catches using trade records from commercial markets. Ecology Letters, 9, 1115–1126.
da Cunha, D. B., da Silva Rodrigues-Filho, L. F., de Luna Sales, J. B. (2017). A review of the mitogenomic phylogeny of the Chondrichthyes. In: da Silva Rodrigues Filho LF, de Luna Sales JB, (Eds.), Chondrichthyes- Multidisciplinary Approach. IntechOpen, pp. 113–126.
de Carvalho, M. R. (1996). Higher-level elasmobranch phylogeny, basal squaleans, and paraphyly. In M. L. J. Stiassny, L. R. Parenti, & G. D. Johnson (Eds.), Interrelationships of fishes (pp. 35–62). Academic Press Inc.
Delsuc, F., Brinkmann, H., & Philippe, H. (2005). Phylogenomics and the reconstruction of the tree of life. Nature Reviews Genetics, 6, 361–375.
Dessimoz, C., & Gil, M. (2010). Phylogenetic assessment of alignments reveals neglected tree signal in gaps. Genome Biology, 11, R37.
Douady, C. J., Dosay, M., Shivji, M. S., & Stanhope, M. J. (2003). Molecular phylogenetic evidence refuting the hypothesis of Batoidea (rays and skates) as derived sharks. Molecular Phylogenetics and Evolution, 26, 215–221.
Dulvy, N. K., Fowler, S. L., Musick, J. A., Cavanagh, R. D., Kyne, P. M., Harrison, L. R., et al. (2014). Extinction risk and conservation of the world’s sharks and rays. eLife, 3, e00590.
Dwivedi, B., & Gadagkar, S. R. (2009). Phylogenetic inference under varying proportions of indel-induced alignment gaps. BMC Evolutionary Biology, 9, 211.
Emms, D. M., & Kelly, S. (2019). OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biology, 20, 238.
Gaitán-Espitia, J. D., Solano-Iguaran, J. J., Tejada-Martinez, D., & Quintero-Galvis, J. F. (2016). Mitogenomics of electric rays: Evolutionary considerations within Torpediniformes (Batoidea; Chondrichthyes). Zoological Journal of the Linnean Society, 178, 257–266.
Haas, B. J., Papanicolaou, A., Yassour, M., Grabherr, M., Blood, P. D., Bowden, J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature Protocols, 8, 1494–1512.
Heinicke,. M. P., Naylor, G. J. P., Hedges, S. B. (2009). Cartilaginous fishes (Chondrichthyes). Pp. 320–327. In: The Timetree of Life. Hedges SB and Kumar S (eds.). Oxford University Press.
Heupel, M. R., Knip, D. M., Simpfendorfer, C. A., & Dulvy, N. K. (2014). Sizing up the ecological role of sharks as predators. Marine Ecology Progress Series, 495, 291–298.
Hughes, L. C., Ortí, G., Huang, Y., Sun, Y., Baldwin, C. C., Thompson, A. W., et al. (2018). Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proceedings of the National Academy of Sciences of the United States of America, 115, 6249–6254.
Human, B. A., Owen, E. P., Compagno, L. J., & Harley, E. H. (2006). Testing morphologically based phylogenetic theories within the cartilaginous fishes with molecular data, with special reference to the catshark family (Chondrichthyes; Scyliorhinidae) and the interrelationships within them. Molecular Phylogenetics and Evolution, 39, 384–391.
Inoue, J. G., Miya, M., Lam, K., Tay, B. H., Danks, J. A., Bell, J., et al. (2010). Evolutionary origin and phylogeny of the modern holocephalans (Chondrichthyes: Chimaeriformes): A mitogenomic perspective. Molecular Biology and Evolution, 27, 2576–2586.
Ioannidis, P., Simao, F. A., Waterhouse, R. M., Manni, M., Seppey, M., Robertson, H. M., et al. (2017). Genomic features of the damselfly Calopteryx splendens representing a sister clade to most insect orders. Genome Biology and Evolution, 9, 415–430.
Irisarri, I., Baurain, D., Brinkmann, H., Delsuc, F., Sire, J. Y., Kupfer, A., et al. (2017). Phylotranscriptomic consolidation of the jawed vertebrate timetree. Nature Ecology and Evolution, 1, 1370–1378.
Kapli, P., Yang, Z., & Telford, M. J. (2020). Phylogenetic tree building in the genomic age. Nature Reviews Genetics, 21, 428–444.
Katoh, K., & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780.
Kousteni, V., Mazzoleni, S., Vasileiadou, K., & Rovatsos, M. (2021). Complete mitochondrial DNA genome of nine species of sharks and rays and their phylogenetic placement among modern Elasmobranchs. Genes, 12, 324.
Kuhnhäuser, B. G., Bellot, S., Couvreur, T. L. P., Dransfield, J., Henderson, A., Schley, R., et al. (2021). A robust phylogenomic framework for the calamoid palms. Molecular Phylogenetics and Evolution, 157, 107067.
Li, C., Matthes-Rosana, K. A., Garcia, M., & Naylor, G. J. (2012). Phylogenetics of Chondrichthyes and the problem of rooting phylogenies with distant outgroups. Molecular Phylogenetics and Evolution, 63, 365–373.
Mallat, J., & Winchell, C. J. (2007). Ribosomal RNA genes and deuterostome phylogeny revisited: more cyclostomes, elasmobranchs, reptiles, and a brittle star. Molecular Phylogenetics and Evolution, 43, 1005–1022.
Matich, P., Heithaus, M. R., & Layman, C. A. (2011). Contrasting patterns of individual specialization and trophic coupling in two marine apex predators. Journal of Animal Ecology, 80, 294–305.
McEachran, J. D., & Aschliman, N. (2004). Phylogeny of batoidea. In J. Carrier, J. Musick, & M. Heithaus (Eds.), Biology of sharks and their relatives (pp. 79–113). CRC Press.
Minh, B. Q., Dang, C. C., Vinh, L. S., & Lanfear, R. (2021). QMaker: Fast and accurate method to estimate empirical models of protein evolution. Systematic Biology, 70, 1046–1060.
Mikula, O., Nicolas, V., Šumbera, R., Konečný, A., Denys, C., Verheyen, E., et al. (2021). Nuclear phylogenomics, but not mitogenomics, resolves the most successful Late Miocene radiation of African mammals (Rodentia: Muridae: Arvicanthini). Molecular Phylogenetics and Evolution, 157, 107069.
Mistry, J., Finn, R. D., Eddy, S. R., Bateman, A., & Punta, M. (2013). Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Research, 41, e121.
Naylor, G. J. P., Caira, J., Jensen, K., Rosana, K., Straube, N., Lakner, C. (2012). Elasmobranch phylogeny: A mitochondrial estimate based on 595 species. In: Carrier J, Musick J, Heithaus M, (Eds.). Biology of sharks and their relatives, 2nd edn. CRC Press, Boca Raton, FL.
Naylor, G. J. P., Ryburn, J. A., Fedrigo, O., & López, J. A. (2005). Phylogenetic relationships among the major lineages of modern elasmobranchs. Science Publishers IncIn W. C. Hamlett & B. G. M. Jamieson (Eds.), Reproductive Biology and Phylogeny (Vol. 3, pp. 1–25). Enfield.
Naylor, G. J. P., Yang, L., Corrigan, S., & de Carvalho, M. R. (2016). Phylogeny and classification of rays. In W. T. White, M. R. de Carvalho, B. Séret, M. F. W. Stehmann, & G. J. P. Naylor (Eds.), Last PR (pp. 10–15). Rays of the world.
Naylor, G. J. P. (2021). Chondrichthyans tree of life. https://sharksrays.org/. Accessed 16 Sep 2021.
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., et al. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution, 32, 268–274.
Oshima, K., & Nishida, H. (2007). Phylogenetic relationships among mycoplasmas based on the whole genomic information. Journal of Molecular Evolution, 65, 249–258.
Parins-Fukuchi, C. (2018). Use of continuous traits can improve morphological phylogenetics. Systematic Biology, 67, 328–339.
Pearce, J., Fraser, M. W., Sequeira, A. M. M., & Kaur, P. (2021). State of shark and ray genomics in an era of extinction. Frontiers in Marine Science, 8, 744986.
Philippe, H., Snell, E. A., Bapteste, E., Lopez, P., Holland, P. W., & Casane, D. (2004). Phylogenomics of eukaryotes: Impact of missing data on large alignments. Molecular Biology and Evolution, 21, 1740–1752.
Roch, S., & Warnow, T. (2015). On the robustness to gene tree estimation error (or lack thereof) of coalescent-based species tree methods. Systematic Biology, 64, 663–676.
Roure, B., Rodriguez-Ezpeleta, N., & Philippe, H. (2007). SCaFoS: A tool for selection, concatenation and fusion of sequences for phylogenomics. BMC Evolutionary Biology, 7, S2.
Seppey, M., Manni, M., & Zdobnov, E. M. (2019). BUSCO: Assessing genome assembly and annotation completeness. Methods in Molecular Biology, 1962, 227–245.
Shen, X. X., Zhou, X., Kominek, J., Kurtzman, C. P., Hittinger, C. T., Rokas, A. (2016). Reconstructing the backbone of the Saccharomycotina yeast phylogeny using genome-scale data. G3 (Bethesda), 6, 3927–3939.
Shirai, S. (1996). Phylogenetic interrelationships of Neoselachians (Chondrichthyes: Euselachii). In M. L. J. Stiassny, L. R. Parenti, & G. D. Johnson (Eds.), Interrelationships of fishes (pp. 9–34). Academic Press Inc.
Stamatakis, A. (2014). RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312–1313.
Stein, R. W., Mull, C. G., Kuhn, T. S., Aschliman, N. C., Davidson, L. N. K., Joy, J. B., et al. (2018). Global priorities for conserving the evolutionary history of sharks, rays and chimaeras. Nature Ecology and Evolution, 2, 288–298.
Uno, Y., Nozu, R., Kiyatake, I., Higashiguchi, N., Sodeyama, S., Murakumo, K., et al. (2020). Cell culture-based karyotyping of orectolobiform sharks for chromosome-scale genome analysis. Communications Biology, 3, 652.
Vélez-Zuazo, X., & Agnarsson, I. (2011). Shark tales: A molecular species-level phylogeny of sharks (Selachimorpha, Chondrichthyes). Molecular Phylogenetics and Evolution, 58, 207–217.
Waterhouse, R. M., Seppey, M., Simão, F. A., Manni, M., Ioannidis, P., Klioutchnikov, G., et al. (2018). BUSCO applications from quality assessments to gene prediction and phylogenomics. Molecular Biology and Evolution, 35, 543–548.
Zhang, N., Zeng, L., Shan, H., & Ma, H. (2012). Highly conserved low-copy nuclear genes as effective markers for phylogenetic analyses in angiosperms. New Phytologist, 195, 923–937.
Acknowledgements
We thank Prof. Tianxing Gao for the kind suggestions.
Funding
This work was supported by Zhejiang Provincial Natural Science Foundation of China (grant number LR21D060003) and National Natural Science Foundation of China (grant number 32200413).
Author information
Authors and Affiliations
Contributions
Z.Q.H. conceived the study. R.R.Z., S.S.C. and P.F.L. collected the data. S.Y.X., R.R.Z., and Z.Q.H. analyzed the data and drafted the manuscript. All authors revised the manuscript and gave final approval for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, S., Zhao, R., Cai, S. et al. Application of genomic markers generated for ray-finned fishes in chondrichthyan Phylogenomics. Org Divers Evol 23, 1005–1012 (2023). https://doi.org/10.1007/s13127-023-00607-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-023-00607-w